Classification Workshop: Introduction to Machine Learning in R with caret
Introduction to Machine Learning in R with caret
Part 1 - What is machine learning? What are the tenets, what is the basic workflow?
Discussion - two questions (5-minutes with the person sitting next to you - then we’ll come together and discuss as a group)
- What is machine learning?
- How is it different than statistics?
Some important things to know and think about:
- Prediction is usually more important than explanation
- Two major types of problems - regression and classification
- Splitting the data to prevent overfitting
Classifcation Problem - Wine varietal identifier
Here is the scenario: we’ve been contacted by a famous vignter in Italy because she suspects that one of the prized varietals (a rare version of Aglianicone that her family has grown for 7 generations) from her vinyard has been stolen, and is being grown and sold to make competitively delicious wine in the United States. The competing winemaker claims that the varietal being grown in the US is from a closely related varietal from the same region, that he obtained legally.
Our customer has hired us to develop an algorithm to determine the likelihood that this is the wine being sold by the competitor was made from the varietal grown on her farm. Unfortunately, we don’t have fancy genomic data to work with, but she has provided us with chemical profiles of a bunch of different wines made from both her grapes and two varietals that the competitor claims to be working with. The owner of the competing US vinyard has graciously provided us with the same type of data from a bunch of his wines to make comparisons on - he’s looking to clear his name (and probably doesn’t also believe that an algorithm can predict whether or not a given wine comes from a certain regional varietal)
Part 2 - Examining the Data
# Getting libraries we need loaded
library(caret)
library(tidyverse)
#Reading in the data from the github repo
wine_train = read_csv(file = "https://raw.githubusercontent.com/keatonwilson/classification_workshop_1/master/data/wine_train.csv")
#https://bit.ly/2xoqHVZ
#
wine_test = read_csv(file = "https://raw.githubusercontent.com/keatonwilson/classification_workshop_1/master/data/wine_test.csv")
#https://bit.ly/2NRXgpp
#Overviews
glimpse(wine_train)## Observations: 168
## Variables: 15
## $ varietal <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1...
## $ alcohol <dbl> 14.23, 13.20, 13.16, 14.37, 13.24, 14.20, 14.3...
## $ malic_acid <dbl> 1.71, 1.78, 2.36, 1.95, 2.59, 1.76, 1.87, 2.15...
## $ ash <dbl> 2.43, 2.14, 2.67, 2.50, 2.87, 2.45, 2.45, 2.61...
## $ alkalinity <dbl> 15.6, 11.2, 18.6, 16.8, 21.0, 15.2, 14.6, 17.6...
## $ magnesium <int> 127, 100, 101, 113, 118, 112, 96, 121, 97, 98,...
## $ total_phenol <dbl> 2.80, 2.65, 2.80, 3.85, 2.80, 3.27, 2.50, 2.60...
## $ flavanoids <dbl> 3.06, 2.76, 3.24, 3.49, 2.69, 3.39, 2.52, 2.51...
## $ nonflav_phenols <dbl> 0.28, 0.26, 0.30, 0.24, 0.39, 0.34, 0.30, 0.31...
## $ proantho <dbl> 2.29, 1.28, 2.81, 2.18, 1.82, 1.97, 1.98, 1.25...
## $ color <dbl> 5.64, 4.38, 5.68, 7.80, 4.32, 6.75, 5.25, 5.05...
## $ hue <dbl> 1.04, 1.05, 1.03, 0.86, 1.04, 1.05, 1.02, 1.06...
## $ OD <dbl> 3.92, 3.40, 3.17, 3.45, 2.93, 2.85, 3.58, 3.58...
## $ proline <int> 1065, 1050, 1185, 1480, 735, 1450, 1290, 1295,...
## $ id <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,...summary(wine_train)## varietal alcohol malic_acid ash
## Min. :1.000 Min. :11.03 Min. :0.740 Min. :1.360
## 1st Qu.:1.000 1st Qu.:12.36 1st Qu.:1.610 1st Qu.:2.210
## Median :2.000 Median :13.05 Median :1.885 Median :2.360
## Mean :1.946 Mean :13.02 Mean :2.369 Mean :2.369
## 3rd Qu.:3.000 3rd Qu.:13.70 3rd Qu.:3.132 3rd Qu.:2.565
## Max. :3.000 Max. :14.83 Max. :5.800 Max. :3.230
## alkalinity magnesium total_phenol flavanoids
## Min. :10.60 Min. : 70.0 Min. :0.980 Min. :0.340
## 1st Qu.:17.07 1st Qu.: 88.0 1st Qu.:1.715 1st Qu.:1.090
## Median :19.50 Median : 98.0 Median :2.335 Median :2.035
## Mean :19.49 Mean : 99.8 Mean :2.282 Mean :1.997
## 3rd Qu.:21.50 3rd Qu.:107.2 3rd Qu.:2.800 3rd Qu.:2.865
## Max. :30.00 Max. :162.0 Max. :3.880 Max. :5.080
## nonflav_phenols proantho color hue
## Min. :0.1400 Min. :0.410 Min. : 1.280 Min. :0.4800
## 1st Qu.:0.2700 1st Qu.:1.235 1st Qu.: 3.147 1st Qu.:0.7800
## Median :0.3400 Median :1.535 Median : 4.850 Median :0.9600
## Mean :0.3664 Mean :1.583 Mean : 5.100 Mean :0.9534
## 3rd Qu.:0.4500 3rd Qu.:1.952 3rd Qu.: 6.263 3rd Qu.:1.1200
## Max. :0.6600 Max. :3.580 Max. :13.000 Max. :1.7100
## OD proline id
## Min. :1.270 Min. : 278.0 Min. : 1.00
## 1st Qu.:1.905 1st Qu.: 500.0 1st Qu.: 45.75
## Median :2.770 Median : 660.0 Median : 90.50
## Mean :2.599 Mean : 745.6 Mean : 90.14
## 3rd Qu.:3.172 3rd Qu.: 996.2 3rd Qu.:136.25
## Max. :4.000 Max. :1547.0 Max. :178.00#Checking for NAs
sum(is.na(wine_train))## [1] 0Ok, so this looks good. We have our item we want to classify in column 1, and all of our features in the rest. For our varietal numbers, 1 and 2 are the local varietals not owned by our customer, but varietal 3 is her special grape. So we’re looking for the presence of any wines made from varietal 3 in the test set.
What do we need to do before we jump into to trying to build some algorithms?
Preprocess! In particular, we need to center and scale the data. caret can do this for us.
Part 3 - Preprocessing
#Setting up the preprocessing algorithm
set.seed(42)
pp = preProcess(wine_train[,-1], method = c("center", "scale"), outcome = wine_train$varietal)
wine_train_pp = predict(pp, wine_train)
wine_train_pp## # A tibble: 168 x 15
## varietal alcohol malic_acid ash alkalinity magnesium total_phenol
## <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 1.48 -0.584 0.220 -1.15 1.88 0.823
## 2 1 0.225 -0.522 -0.831 -2.44 0.0140 0.585
## 3 1 0.176 -0.00786 1.09 -0.264 0.0830 0.823
## 4 1 1.65 -0.371 0.474 -0.794 0.912 2.49
## 5 1 0.274 0.196 1.82 0.444 1.26 0.823
## 6 1 1.45 -0.540 0.293 -1.27 0.843 1.57
## 7 1 1.68 -0.442 0.293 -1.44 -0.262 0.346
## 8 1 1.28 -0.194 0.873 -0.558 1.46 0.505
## 9 1 2.22 -0.646 -0.722 -1.62 -0.193 0.823
## 10 1 1.03 -0.903 -0.360 -1.03 -0.124 1.11
## # ... with 158 more rows, and 8 more variables: flavanoids <dbl>,
## # nonflav_phenols <dbl>, proantho <dbl>, color <dbl>, hue <dbl>,
## # OD <dbl>, proline <dbl>, id <dbl>#We also need to add this same processing algorithm to the test data.
wine_test_pp = predict(pp, wine_test)
#We also need to make the varietal category a factor in both datasets
wine_train_pp = wine_train_pp %>%
mutate(varietal = factor(varietal))Part 4 - Model Testing and Tuning
There are a ton of classification models to choose from - when starting ML stuff, this can be a really daunting part of the thing. Today, we’re going to explore a couple of bread-and-butter models:
1. k-nn - nearest neighbord classifier
2. Naive Bayes
3. Decision Trees
4. Support Vector Machines
I’m not going to go into the math of how all of these operate at all. It’s the beyond the scope of this workshop, but here is a good overview: https://medium.com/@sifium/machine-learning-types-of-classification-9497bd4f2e14
One thing that we need to talk about briefly is resampling - this is the method we’re going to use to assess how ‘good’ a model is, without applying it to the test data. There are a couple of main ways to do this:
1. bootstrapping - random sampling within the dataset with replacement. Pulling a bunch of subsets of the data and looking at how the model performs across these subsets.
2. Repeated n-fold cross-validation - does a bunch of splitting into training and test data within the training set, and then averages accuracy or RMSE across all these little mini-sets.
We’re going to use the second type.
# setting up the control object to feed to all of the subsequent models
fit_control = trainControl(method = "repeatedcv", number = 5, repeats = 5)
#models
#knn
knn_model = train(varietal ~ ., data = wine_train_pp,
method = "knn", trControl = fit_control)
#naive bays
bayes_model = train(varietal ~ ., data = wine_train_pp,
method = "nb", trControl = fit_control)
#CART
cart_model = train(varietal ~ ., data = wine_train_pp,
method = "rpart", trControl = fit_control)
#svm
svm_model = train(varietal ~ ., data = wine_train_pp,
method = "svmRadial", trControl = fit_control)The models default to using accuracy as the score to determine how good they are. Accuracy is the percentage of the predictions made by the model that are correct.
Let’s comapre models
results = resamples(list(knn = knn_model, bayes = bayes_model, cart = cart_model, svm = svm_model))
summary(results)##
## Call:
## summary.resamples(object = results)
##
## Models: knn, bayes, cart, svm
## Number of resamples: 25
##
## Accuracy
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## knn 0.9117647 0.969697 0.9714286 0.9761793 1 1 0
## bayes 0.9696970 1.000000 1.0000000 0.9964349 1 1 0
## cart 0.9696970 0.969697 1.0000000 0.9867380 1 1 0
## svm 0.9696970 1.000000 1.0000000 0.9952228 1 1 0
##
## Kappa
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## knn 0.8680466 0.9537815 0.9569496 0.9641544 1 1 0
## bayes 0.9539106 1.0000000 1.0000000 0.9945892 1 1 0
## cart 0.9539106 0.9542936 1.0000000 0.9799294 1 1 0
## svm 0.9542936 1.0000000 1.0000000 0.9927833 1 1 0dotplot(results)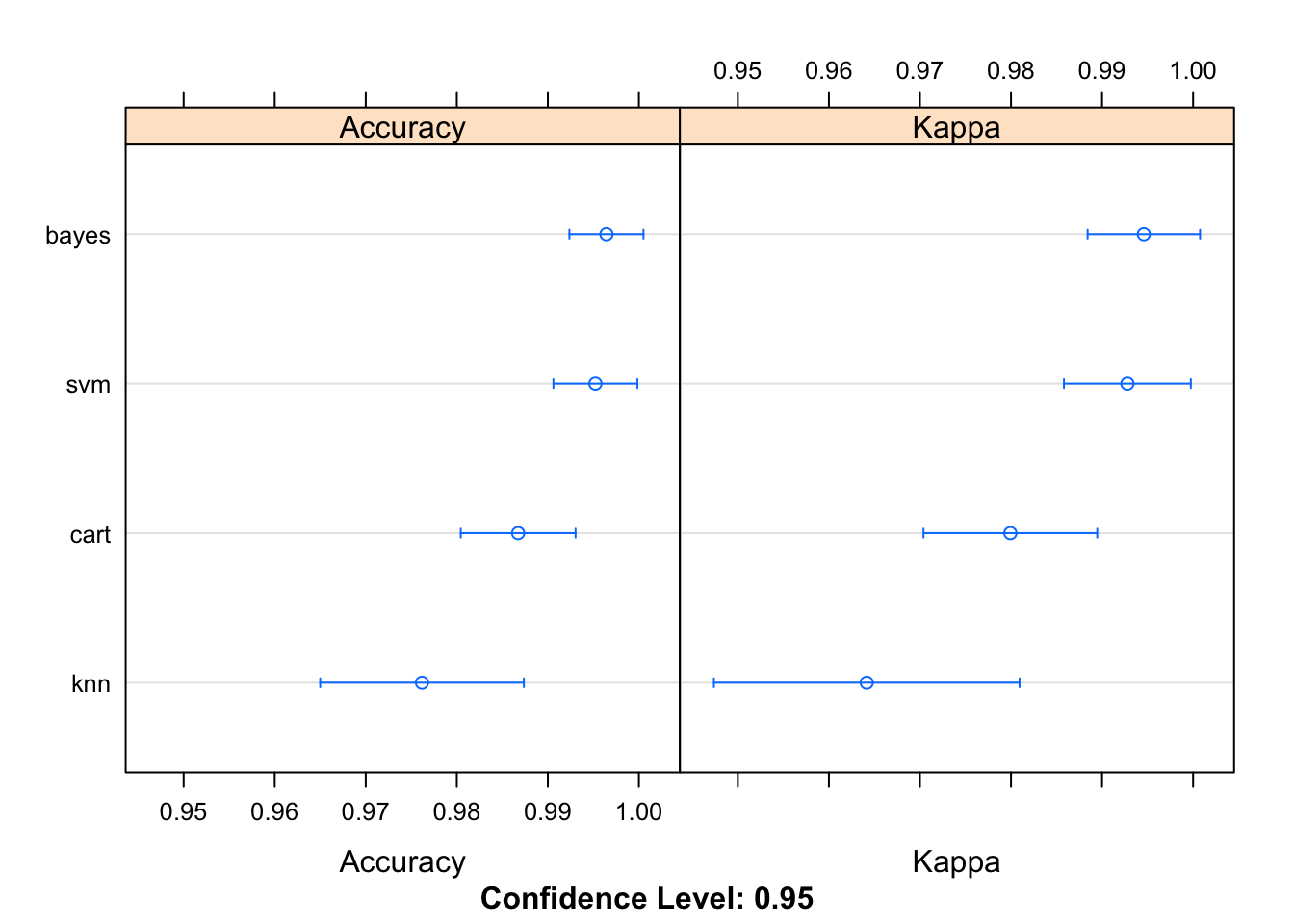
svm_model## Support Vector Machines with Radial Basis Function Kernel
##
## 168 samples
## 14 predictor
## 3 classes: '1', '2', '3'
##
## No pre-processing
## Resampling: Cross-Validated (5 fold, repeated 5 times)
## Summary of sample sizes: 135, 134, 133, 135, 135, 134, ...
## Resampling results across tuning parameters:
##
## C Accuracy Kappa
## 0.25 0.9893028 0.9838228
## 0.50 0.9904792 0.9855912
## 1.00 0.9952228 0.9927833
##
## Tuning parameter 'sigma' was held constant at a value of 0.05224556
## Accuracy was used to select the optimal model using the largest value.
## The final values used for the model were sigma = 0.05224556 and C = 1.We can also look at the relative amount of false negatives and false positives with a confusion matrix.
predictions = predict(svm_model, wine_train_pp)
confusionMatrix(predictions, wine_train_pp$varietal)## Confusion Matrix and Statistics
##
## Reference
## Prediction 1 2 3
## 1 56 0 0
## 2 0 65 0
## 3 0 0 47
##
## Overall Statistics
##
## Accuracy : 1
## 95% CI : (0.9783, 1)
## No Information Rate : 0.3869
## P-Value [Acc > NIR] : < 2.2e-16
##
## Kappa : 1
## Mcnemar's Test P-Value : NA
##
## Statistics by Class:
##
## Class: 1 Class: 2 Class: 3
## Sensitivity 1.0000 1.0000 1.0000
## Specificity 1.0000 1.0000 1.0000
## Pos Pred Value 1.0000 1.0000 1.0000
## Neg Pred Value 1.0000 1.0000 1.0000
## Prevalence 0.3333 0.3869 0.2798
## Detection Rate 0.3333 0.3869 0.2798
## Detection Prevalence 0.3333 0.3869 0.2798
## Balanced Accuracy 1.0000 1.0000 1.0000Not particularly informative, given the model did a 100% accurate job of predicting on the training-data, but you get the gist.
Part 5 - Using the model on the test data.
wine_test_pp$pred = predict(svm_model, wine_test_pp)
#What is your conclusion?
wine_test_pp$pred## [1] 1 1 2 1 3 2 2 2 2 2
## Levels: 1 2 3Part 6 - Continuing Practice
Some resources if you want to get better at this:
1. Kaggle - an online community of data scientists - lots of cool datasets to play with, and competitions!
2. www.datacamp.com - great series of lessons on machine learning, including classification and regression, with way deeper dives on the power of caret and other packages (python too!)
3. Machine Learning with R - Brett Lantz. Great book!
